powerfulpictures – Venturing into Forax
 Colorful monochrome imaging of SHO data
Read More ›
Colorful monochrome imaging of SHO data
Read More ›
powerfulpictures – Himarari-8 watching over us: one year compressed into 16min
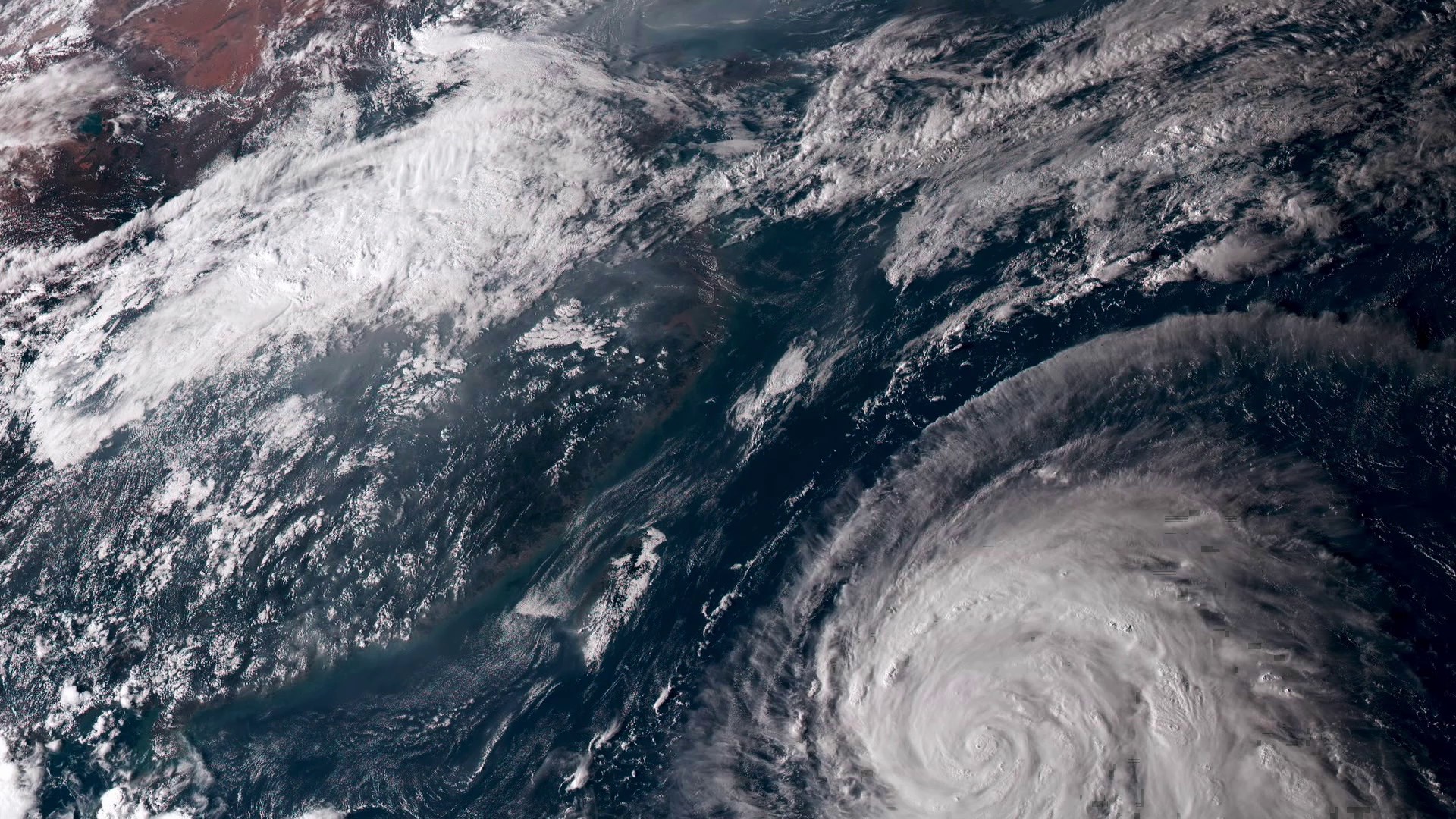 From 20,000 miles up, our home planet is a hypnotic swirl of the familiar and the sublime
Read More ›
From 20,000 miles up, our home planet is a hypnotic swirl of the familiar and the sublime
Read More ›
powerfulpictures – 23 hours
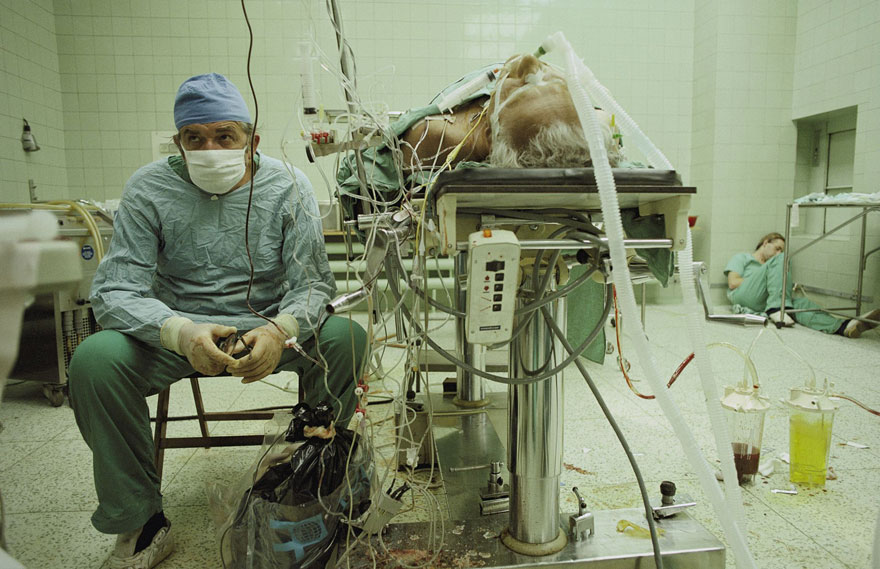 about a surgeon who for the first time performed a heart transplant
Read More ›
about a surgeon who for the first time performed a heart transplant
Read More ›